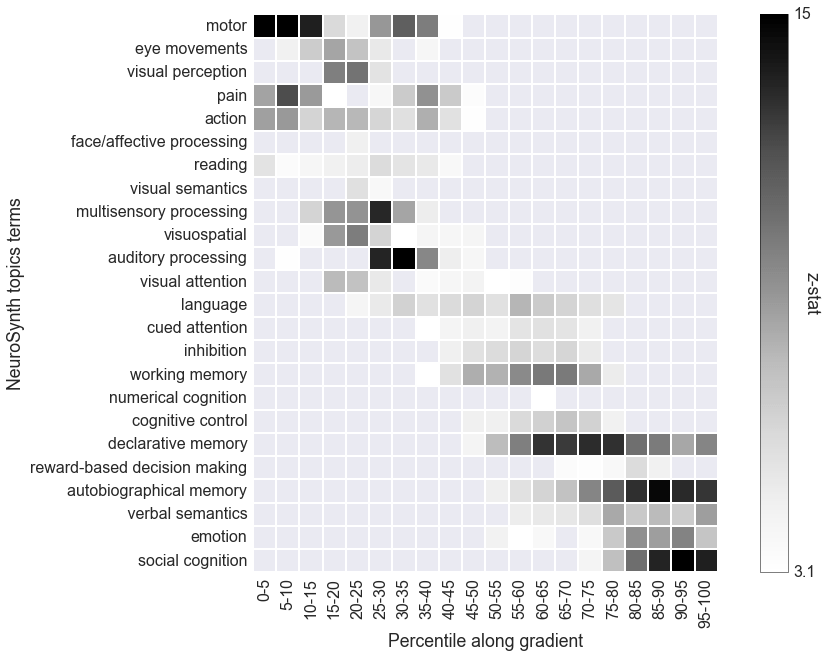
Meta-analysis using NeuroSynth
-
PDF | SI | Link | Code | | Data | Margulies DS, Ghosh SS, Goulas A, Falkiewicz M, Huntenburg JM, Langs G, Bezgin G, Eickhoff SB, Castellanos FX, Petrides M, Jefferies E, Smallwood J (2016) Situating the default-mode network along a principal gradient of macroscale cortical organization Proc Natl Acad Sci U S A 113(44): 12574–12579
% matplotlib inline
from neurosynth.base.dataset import Dataset
from neurosynth.analysis import decode
import pandas as pd
import numpy as np
import seaborn as sns
import matplotlib as mpl
import matplotlib.pyplot as plt
mpl.rcParams['svg.fonttype'] = 'none'
def getOrder(d, thr):
dh = []
for i in range(0,len(d)):
di = d[i]
dh.append(np.average(np.array(xrange(0,len(d[i]))) + 1, weights=di))
heatmapOrder = np.argsort(dh)
return heatmapOrder
# Import neurosynth database:
pickled_dataset = '../topography/metaanalysis/neurosynth/dataset.pkl' #'gradient_data/neurosynth/dataset.pkl'
dataset = Dataset.load(pickled_dataset)
%%bash
# Create masks for metaanalysis
for i in `seq 0 25`; do
let ind="${i} + 1"
wb_command -cifti-merge gradient_data/embedded/ciftis/hcp.embed.${i}.dscalar.nii \
-cifti gradient_data/embedded/ciftis/hcp.embed.dscalar.nii -column ${ind}
wb_command -cifti-separate gradient_data/embedded/ciftis/hcp.embed.${i}.dscalar.nii COLUMN \
-metric CORTEX_LEFT gradient_data/embedded/ciftis/hcp.embed.${i}.L.metric \
-metric CORTEX_RIGHT gradient_data/embedded/ciftis/hcp.embed.${i}.R.metric \
-volume-all gradient_data/embedded/ciftis/hcp.embed.${i}.volume.nii
wb_command -metric-to-volume-mapping gradient_data/embedded/ciftis/hcp.embed.${i}.L.metric \
gradient_data/templates/S900.L.midthickness_MSMAll.32k_fs_LR.surf.gii \
gradient_data/templates/MNI152_T1_2mm_brain.nii.gz gradient_data/embedded/volumes/volume.${i}.L.nii \
-ribbon-constrained gradient_data/templates/S900.L.white_MSMAll.32k_fs_LR.surf.gii \
gradient_data/templates/S900.L.pial_MSMAll.32k_fs_LR.surf.gii
wb_command -metric-to-volume-mapping gradient_data/embedded/ciftis/hcp.embed.${i}.R.metric \
gradient_data/templates/S900.R.midthickness_MSMAll.32k_fs_LR.surf.gii \
gradient_data/templates/MNI152_T1_2mm_brain.nii.gz gradient_data/embedded/volumes/volume.${i}.R.nii \
-ribbon-constrained gradient_data/templates/S900.R.white_MSMAll.32k_fs_LR.surf.gii \
gradient_data/templates/S900.R.pial_MSMAll.32k_fs_LR.surf.gii
# combine:
fslmaths gradient_data/embedded/volumes/volume.${i}.L.nii \
-add gradient_data/embedded/volumes/volume.${i}.R.nii \
-add gradient_data/embedded/ciftis/hcp.embed.${i}.volume.nii \
gradient_data/embedded/volumes/volume.orig.${i}.nii
fslmaths gradient_data/embedded/volumes/volume.${i}.L.nii \
-add gradient_data/embedded/volumes/volume.${i}.R.nii \
-add gradient_data/embedded/ciftis/hcp.embed.${i}.volume.nii \
-abs -bin gradient_data/embedded/volumes/mask.${i}.nii
p=`fslstats gradient_data/embedded/volumes/volume.orig.${i}.nii -R | awk '{print $1;}'`
fslmaths gradient_data/embedded/volumes/mask.${i}.nii -mul ${p#-} gradient_data/embedded/volumes/mask.${i}.nii
fslmaths gradient_data/embedded/volumes/volume.orig.${i}.nii.gz \
-add gradient_data/embedded/volumes/mask.${i}.nii \
gradient_data/embedded/volumes/volume.${i}.nii.gz
# extract masks:
mkdir gradient_data/embedded/volumes/emb_masks_${i}
for j in `seq 0 5 95`; do
let k="${j} + 5"
fslmaths gradient_data/embedded/volumes/volume.${i}.nii \
-thr `fslstats gradient_data/embedded/volumes/volume.${i}.nii -P ${j}` \
-uthr `fslstats gradient_data/embedded/volumes/volume.${i}.nii -P ${k}` \
-bin gradient_data/embedded/volumes/emb_masks_${i}/volume_$(printf %02d $j)_$(printf %02d $k).nii
done
done
# Copy masks from Gradient_0 to masks directory
cp gradient_data/embedded/volumes/emb_masks_0/volume* gradient_data/masks/
rm -f gradient_data/embedded/volumes/vol*L.nii
rm -f gradient_data/embedded/volumes/vol*R.nii
rm -f gradient_data/embedded/volumes/mask*.nii.gz
rm -f gradient_data/embedded/ciftis/hcp.embed.*.L.metric
rm -f gradient_data/embedded/ciftis/hcp.embed.*.R.metric
# Analysis with 24 terms:
features = pd.read_csv('gradient_data/neurosynth/v3-topics-50.txt', sep='\t', index_col=0)
topics_to_keep = [ 1, 4, 6, 14,
18, 19, 23, 25,
20, 21, 27, 29,
30, 31, 33, 35,
36, 38, 37, 41,
44, 45, 48, 49]
labels = ['face/affective processing', ' verbal semantics', 'cued attention', 'working memory',
'autobiographical memory', 'reading', 'inhibition', 'motor',
'visual perception', 'numerical cognition', 'reward-based decision making', 'visual attention',
'multisensory processing', 'visuospatial','eye movements', 'action',
'auditory processing', 'pain', 'language', 'declarative memory',
'visual semantics', 'emotion', 'cognitive control', 'social cognition']
features = features.iloc[:, topics_to_keep]
features.columns = labels
dataset.add_features(features, append=False)
# removed_as_noise = [0,5,9,12,17,40] # from 30 terms that were above threshold
# labels_noise = ['resting-state', 'dementia', 'development', 'misc', 'task timing', 'lateralization']
# Gradient 1
decoder = decode.Decoder(dataset, method='roi')
# Set threshold:
thr = 3.1
vmin = 0
vmax = 15
tot = 5
data = decoder.decode([str('gradient_data/masks/volume_%02d_%02d.nii.gz' % (i * tot, (i * tot) + tot))
for i in xrange(0,100/tot)])
df = []
df = data.copy()
newnames = []
[newnames.append(('%s-%s' % (str(i * tot), str((i*tot) + tot)))) for i in xrange(0,len(df.columns))]
df.columns = newnames
df[df<thr] = 0
heatmapOrder = getOrder(np.array(df), thr)
sns.set(context="paper", font="sans-serif", font_scale=2)
f, (ax1) = plt.subplots(nrows=1,ncols=1,figsize=(15, 10), sharey=True)
plotData = df.reindex(df.index[heatmapOrder])
cax = sns.heatmap(plotData, linewidths=1, square=True, cmap='Greys', robust=False,
ax=ax1, vmin=thr, vmax=vmax, mask=plotData == 0)
sns.axlabel('Percentile along gradient', 'NeuroSynth topics terms')
cbar = cax.collections[0].colorbar
cbar.set_label('z-stat', rotation=270)
cbar.set_ticks(ticks=[thr,vmax])
cbar.set_ticklabels(ticklabels=[thr,vmax])
cbar.outline.set_edgecolor('black')
cbar.outline.set_linewidth(0.5)
plt.draw()
f.savefig('gradient_data/figures/fig.neurosynth.svg', format='svg')